Genome Architecture Mapping (GAM)
Keywords
Chromatin structure, three-dimensional chromatin topology, long-range gene regulation, nuclear sections, locus co-segregation, chromosome conformation capture, 3C, enhancers, super-enhancers, transcritption factors, new targets
Invention Novelty
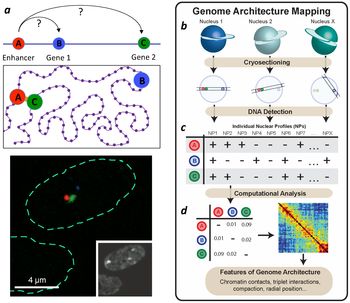
Genome Architecture Mapping (GAM) is a novel genome-wide method for measuring three-dimensional chromatin topology without ligation, that avoids tissue dissociation and enables direct targeting of specific cells, or diseased states, within a tissue. GAM detects a previously inaccessible complexity in 3D genome folding and its roles in gene regulation, opening new avenues for the functional and mechanistic interpretation of genome sequence and discovery of novel targets that drive complex genetic disease.
Value Proposition
GAM is a powerful approach to map 3D genome conformation, especially multi-way contacts between regulatory regions and target genes. GAM is ideally suited to the analysis of specific cell types within tissue samples, without dissociation, requiring minute samples. It reproduces general features of chromatin organization also uncovered by 3C-based methods, but it more powerful in detecting contacts that span large genomic distances (>1 Mb), important to discover the genes affected by disease-associated, non-coding mutations. GAM uniquely reveals multiway contacts between super-enhancers, discovers decondensation events at long neuronal genes, many of which producing many protein isoforms, and associated with complex diseases. GAM is inherently a spatial-omics technology, and its ongoing developments add parallel quantification of the spatial transcriptome and proteome. GAM developments address the interpretability of complex genomes towards identification of new targets for therapy.
Technology Description
GAM extracts spatial information by sequencing DNA from a large collection of thin nuclear sections and quantifying the frequency of locus co-segregation.
Commercial Opportunity
We are looking for industrial partners to cooperate on optimizing and automating GAM technology as well as on validating it in cooperation projects of interest.
Development Status
First version of GAM established; proof of concept demonstrated in complex tissue; in-house whole genome amplification pipeline developed; expansion of GAM into multimodal technologies (RNA-GAM, protein-GAM) ongoing.
Patent Situation
Patents granted in EP (BE, CH, DE, DK, FR, GB, NL, SE), CN, IL, JP, US; pending in CA
Further Reading
Nature. doi:10.1038/nature21411; R.A. Beagrie, A. Scialdone et al. (2017)
Nature. doi.org/10.1038/s41586-021-04081-2; W. Winick-Ng, A. Kukalev et al. (2021)
BioRxiv. doi.org/10.1101/2020.07.31.230284, Beagrie et al. (2021)